PRMT5 and Cancer
William Pratt, CH3 Biosystems Lab Intern
Protein methylation is a common post translational modification that has grown in importance as a research topic in recent years. The role of protein methylation in cancer is extremely complicated and thus very difficult to discuss broadly. This series of posts focuses on the relationship between protein arginine methyltransferases (specifically PRMT5) and cancer (specifically colorectal and liver cancer). Two studies, both by Zhang et al., have recently shown that PRMT5 pathways can be manipulated (through knockdown via PRMT5 specific siRNA or through inhibition via AMI-1) to inhibit CRC and HCC tumor proliferation and migration. Additionally, drug-targeting of PRMT5 has been shown to reduce tumor mass by 65.1% to 75.5%.
First, some background information on protein arginine methylation. Post translational modifications (PTMs) refer to the altering of proteins after they have been synthesized, and are important for proper cell signaling. PTMs can occur through a variety of mechanisms (phosphorylation, acetylation, N-linked glycosylation, amidation, hydroxylation, methylation, etc.) and effectively widens the chemical repertoire of proteins (1). Methylation is the seventh most common PTM, with over 1500 instances recorded and growing.
Methylation is the most basic form of alkylation, and refers to the substitution of a hydrogen atom with a methyl (CH3) group. Enzymes called methyltransferases add methyl groups to side chains of no less than eight of the twenty amino acids, most commonly on nitrogen or oxygen atoms, but also on carbon atoms (2). The methylation of arginine side chains will be our focus in these web posts because these proteins have been found to be significant in a number of cancers.
The addition of methyl groups does not affect the charge on the residue, but it does increase the region’s bulkiness and hydrophobicity. Methylated proteins’ interactions with their binding partners can thus be affected by this change, impacting the physiological functions of the modified protein. Proteins affected by PRMTs include histones, splicing factors, transcription factors, and nucleic acid binding factors.
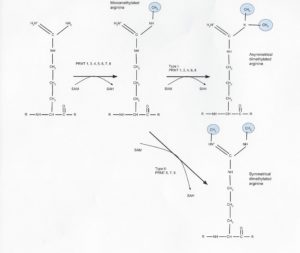
There are nine know protein arginine methyltransferases (PRMTs), and they all use s-adenosyl methionine (SAM) to add methyl groups to the nitrogen atoms of the arginine side chain in several different configurations (Fig. 1). PRMT type I (PRMT1, 3, 4, 6, and 8) and II (5,7, and 9) are responsible for this process in humans, both being capable of producing the mono methylated form and with type I and II also producing asymmetrical and symmetrical dimethylated forms, respectively (3). This process was long thought to be irreversible, but in 2007 the first (and so far, only) protein demethylase (JMJD6) was discovered (4).
Figure 1: PRMT type and methylation locations.
Cancer
The term cancer refers to a group of diseases that are characterized by uninhibited cell growth with the potential for forming malignant tumors. After an initial tumor has formed, the disease can spread to other parts of the body. Globally cancer affects over 90 million people (6), with a five year survival rate of 66% (7).
The majority of cancers are caused by environmental factors, with less than 10% attributed to heritable genetic factors (8). Environmental factors (chemicals, diet and exercise, radiation, infection, etc.) cause cancer by changing the genes in cells. In most cases there must be multiple mutations to result in tumor formation. All cancer cells show the six hallmarks of cancer (9), which must be present in order for a malignant tumor to form. These hallmarks are: Cell growth and division without positive feedback, cell growth and division despite negative feedback, avoidance of apoptosis, avoidance of senescence, blood vessel incorporation, and invasion of healthy tissue.
Some of the most important regulators in cancer are tumor suppressor genes and oncogenes. Tumor suppressor genes effectively protect cells from cancer by regulating the cell cycle or promoting apoptosis. Oncogenes promote cell longevity, and if mutated can prevent cells from undergoing apoptosis.
NEXT TIME: THE INTERSECTION OF CANCER AND PROTEIN METHYLATION (2 OF 3)
References
- Khoury, George A.; Baliban, Richard C. & Christodoulos A. Floudas, Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Scientific Reports 1.90, 90. (2011).
- Walsh CT, Posttranslational Modifications of Proteins: Expanding Nature’s Inventory, Greenwood Village, CO, Roberts and Company Publishers , (2006).
- Aletta and J. Hu, Protein arginine methylation in health and disease, Biotechnology Annual Review , 203-224(2008).
- Poulard, L. Corbo and M. Le Romancer, Protein arginine methylation/demethylation and cancer, Oncotarget 7.41 ,67532-67550 (2016).
- Aletta, T. Cimato and M. Ettinger, Protein methylation: a signal event in post-translational modification, Trends in Biochemical Sciences 23.3, 89-91(1998).
- Vos T et al. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015 , Lancet 388.10053, 1545–1602 (2016)
- SEER Stat Fact Sheets: All Cancer Sites . National Cancer Institute , (2014).
- Anand P, Kunnumakkara AB, Kunnumakara AB, Sundaram C, Harikumar KB, Tharakan ST, Lai OS, Sung B, Aggarwal BB, Cancer is a preventable disease that requires major lifestyle changes . Pharmaceutical Research . 9, 2097–116. (2008)
- Hanahan D, Weinberg RA, The hallmarks of cancer. Cell , 1, 57–70. (2000).
- Bedford and S. Clarke, Protein Arginine Methylation in Mammals: Who, What, and Why, Molecular Cell , 33.1, 1-13, (2009).
- Hou, H. Peng, K. Ayyanathan, K.P. Yan, E.M. Langer, G.D. Longmore, F.J. RauscherIII, The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression Mol. Cell. Biol ., 28, 3198-3207 (2008).
- Pal, S.N. Vishwanath, H. Erdjument-Bromage, P. Tempst, S. SifHuman, SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes Mol. Cell. Biol., 24 , 9630-9645 (2004).
- FM Boisvert et al., Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev 19 , 671–676 (2005).
- MM Adams et al., 53BP1 oligomerization is independent of its methylation by PRMT1. Cell Cycle 4 , 1854–1861 (2005).
- El-Andaloussi et al., Arginine methylation regulates DNA polymerase beta. Mol Cell 22 51–62 (2006).
- Sgarra,The AT-hook of the chromatin architectural transcription factor high mobility group A1a is arginine-methylated by protein arginine methyltransferase 6. J Biol Chem 281 , 3764–3772 (2006).
- Cheung, L.C. Chan, A. Thompson, M.L. Cleary, C.W. So Protein arginine-methyltransferase-dependent oncogenesis Nat. Cell Biol . 9 , 1208-1215 (2007).
- Zhang, S. Dong, R. Zhu, C. Hu, J. Hou, Y. Li, Q. Zhao, X. Shao, Q. Bu, H. Li, Y. Wu, X. Cen and Y. Zhao, Targeting protein arginine methyltransferase 5 inhibits colorectal cancer growth by decreasing arginine methylation of eIF4E and FGFR3, Oncotarget , 6.26 , 22799-22811, (2015).
- Zhang, S. Dong, Z. Li, L. Lu, S. Zhang, X. Chen, X. Cen and Y. Wu, Targeting protein arginine methyltransferase 5 inhibits human hepatocellular carcinoma growth via the downregulation of beta-catenin, Journal of Translational Medicine , 13.1 , (2015).